Data Searches
To access the different data searches, click on 'search' in the menu header and then select the data type you would like to search. To learn more about each search interface, please see the links below the figure.

Search Colleagues
Search Colleague can be accessed through the Search menu in the header. It is a page allows for browsing a list of registered CottonGen users for collaboration and assistance on Cottongen-related topics.
CottonGen Colleague Search Interface.

Search Genes and Transcripts
[ Video Tutorial: How to Search for Genes and Transcripts ]
Search Genes and Transcripts can be accessed through the Search menu in the header. It is a page where users can search for genes and transcripts.
from various datasets available in CottonGen. Users can search for genes from various datasets: predicted genes from whole genome assemblies, a single non-redundant list of cotton genes with gene symbols (Cotton Gene Database), or gene and mRNA sequences parsed out from the NCBI nucleotide database. These genes and mRNAs parsed out from the NCBI sequences are aligned to the reference whole genome sequences when possible. When expert-contributed information is available, these gene names are associated directly with the predicted genes from the whole genome assembly. Users can also search for transcripts from RefTrans sets or reference transcriptome sets built from all publicly available transcripts. For more details, refer to the 'Description of Gene and Transcript Dataset' page.
The search interface of Search Gene and Transcripts. The Query session is on the left and the Downloadable Fields is on the right. See detailed explanation at below.
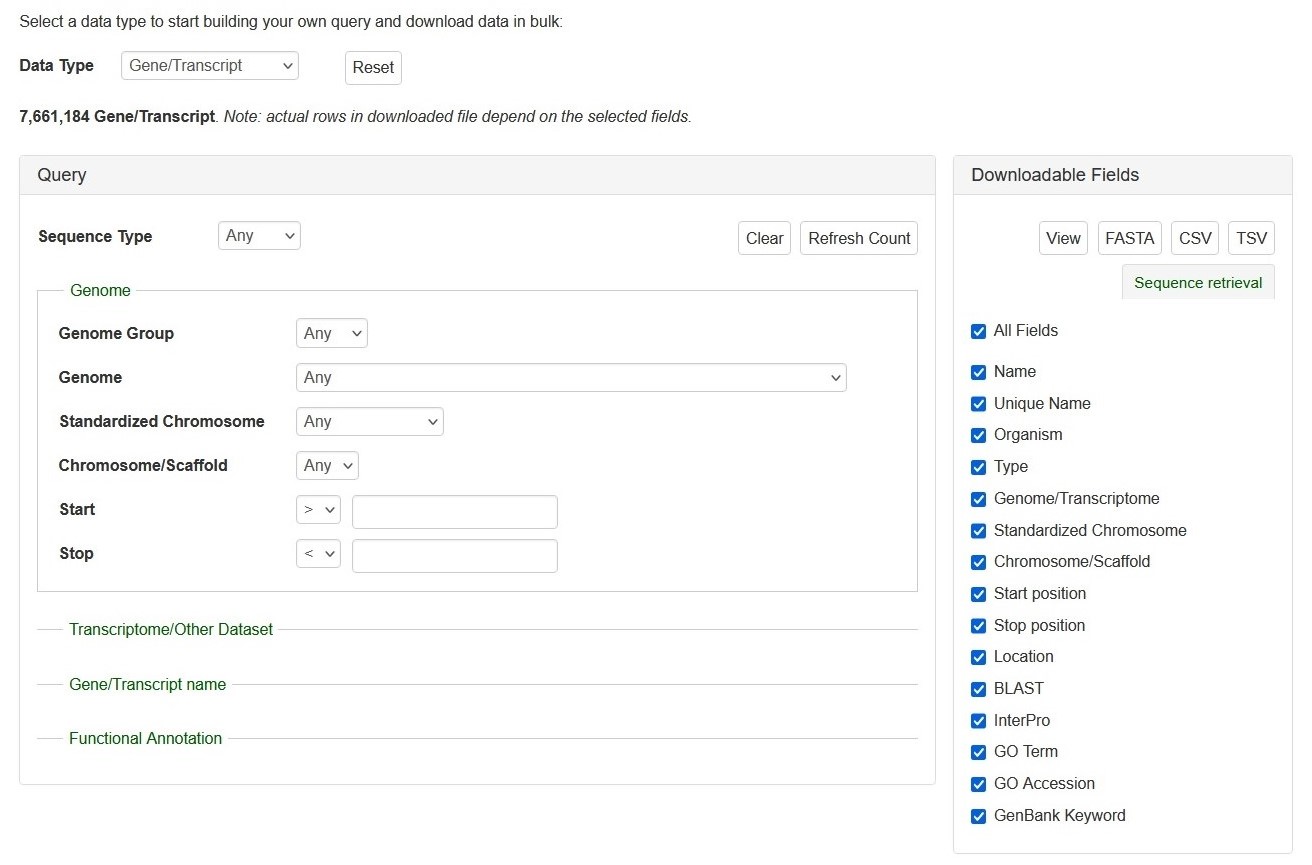
Search Genes and Transcripts can be limited to datasets by four categories (red arrows on left), such as genome and RefTrans assemblies or NCBI genes, by giving gene or transcript names, or by functional annotations. Here are the descriptions of how to search Gene/Transcript data:
There are left (Query) and right (downloadable fields) sessions in the search interface.
Query (Please Note: All the search categories below, except the file upload, can be combined.)
1. Genome
- Genome Group: Users can limit their results of predicted genes by genome groups (such as AD, A, D, etc).
- When a genome group is chosen in the drop-down menu next to 'Genome Group':
- The corresponding genomes are dynamically displayed in the 'Standardized Chromosome'
- The corresponding genomes are dynamically displayed in the 'Genome' and
- When a genome group is chosen in the drop-down menu next to 'Genome Group':
- Genome and Chromosome/Scaffold: Users can limit their results of predicted genes by their genome location.
- When a genome assembly is chosen in the drop-down menu next to 'Genome', the corresponding chromosome or scaffold names are dynamically displayed in the 'Chromosome/Scaffold'. Choose any option and then type in the position in bp in the text boxes.
- Standardized Chromosome: The standardized chromosome names (given by CottoGen) for each genome group.
- When a genome group name is chosen in the drop-down list next to 'Genome Group' and choose an option from the 'Standardized Chromosome' list:
- The search result contains genes or transcripts on the same standard chromosome from all genomes in the genome group.
- When a genome group name is chosen in the drop-down list next to 'Genome Group' and choose an option from the 'Standardized Chromosome' list:
- Start and Stop: Users can limit their results of predicted genes by their genome location.
- When a genome assembly is chosen in the drop-down menu next to 'Dataset', the corresponding chromosome or scaffold names are dynamically displayed in the 'Genome Location' drop-down menu. Choose any option and then type in the position in bp in the text boxes.
2. Transcriptome/Other Dataset: Use this drop-down menu to limit the results to sequences from a specific dataset.
3. Gene/Transcript Name: Users can search genes and transcripts by name for an exact match, that contains, starts with, or ends with the input, by selecting the desired option from the drop-down menu. The search is case-insensitive.
4. Functional Annotation: Users can limit their results by associated functional terms. Predicted genes from whole genome assembly and transcripts have been annotated with some of the following: homology to genes of closely related or plant model species, InterPro protein domains, GO terms, KEGG pathway, and ortholog terms. Users can enter any protein name (eg. polygalacturonase), KEGG term/EC number (eg. resistance, EC:1.4.1.3), GO term (eg. cell cycle, ATP binding), or InterPro term (eg. zinc finger) in the text box to limit the results with the entries that are associated with the functional annotation terms.
Downloadable Fields
1. View|FASTA|CSV|TSV: Users can 'View' the query results on the web interface or download the results in the format of 'FASTA' (of the gene/transcript sequences) or CSV (Comma Separated Values) or TSV (Tab-Separated Values).
2. The All Fields List: A list of fields that users can decide whether in the search results to view or download.
If you have any questions/comments/feedback about this search page, please let us know via the contact form.
Search Genotypes
Search SNP Genotype can be accessed through the Search menu in the header. It is a page with SNP as the default tab followed by the SSR tab. Users can search SNP genotype data by dataset name, marker name, germplasm name, and/or species.
Below is the web interface of Search SNP Genotype with examples of selected Dataset, Species, Germplasm Name, and part of the search results.

Please Note: All the search categories below can be combined and setting multiple filters will only return results matching all criteria.
1. Dataset: Users can search SNP genotype data by dataset from a dropdown list.
2. Species: Users can search for the SNP genotype dataset from a specific species by choosing from the drop-down menu. Multiple options can be selected by holding down the "Ctrl" key.
3. Germplasm Name: Users can search for an SNP genotype dataset from a specific germplasm by choosing from the drop-down menu. Multiple options can be selected by holding down the "Ctrl" key.
4. SNP: Users can search for an SNP genotype dataset that uses a specific marker by typing the marker name in the text box. Users can search for an exact match, that contains, starts with, or ends with the input, by selecting the desired option from the drop-down menu.
5. Genome: Users can search SNP genotype data by the anchored position of SNPs in one of the whole genome sequences. Choose a genome in the drop-down menu next to 'Genome' then the corresponding chromosome or scaffold names will be dynamically generated in the 'Chr/Scaffold' drop-down menu. Choose any option and then type in the position in bp in the text boxes.
6. Search Result: Results are returned in a table that can be downloaded from the entire table or the table with polymorphism data only.
Click on the SSR tab to start an SSR Genotype search.
--- will be completed soon
If you have any questions/comments/feedback about this search page, please let us know via the contact form.
Search Germplasm
The Germplasm Search can be accessed through the Search menu in the header. Information about germplasms linked to data in CottonGen can be searched by name, species, type, collection, pedigree, country of origin, or image. A list of germplasm names, in a text file, can also be uploaded to search for multiple germplasms at once.

If you have any questions/comments/feedback about this search page, please let us know via the contact form.
Publications
Search Publications can be accessed through the Search menu in the header. It is a page where users can search for publications using a combination of keywords (in the abstract or title), all or partial titles, authors, and other categories. Search results link to the publication detail pages that contain the abstract, citation, external link to the full article, and other details. The CottonGen houses information about publications on cotton genomics, genetics, and breeding research. Details about publications were imported to the CottonGen from NCBI PubMed and the USDA National Agricultural Library using the query: (abstract: trait OR QTL OR gene OR genome OR map OR microsatellite OR annotation OR EST OR marker OR sequence) AND (abstract: cotton). Additionally, details of publications from other journals not present in these databases are added.
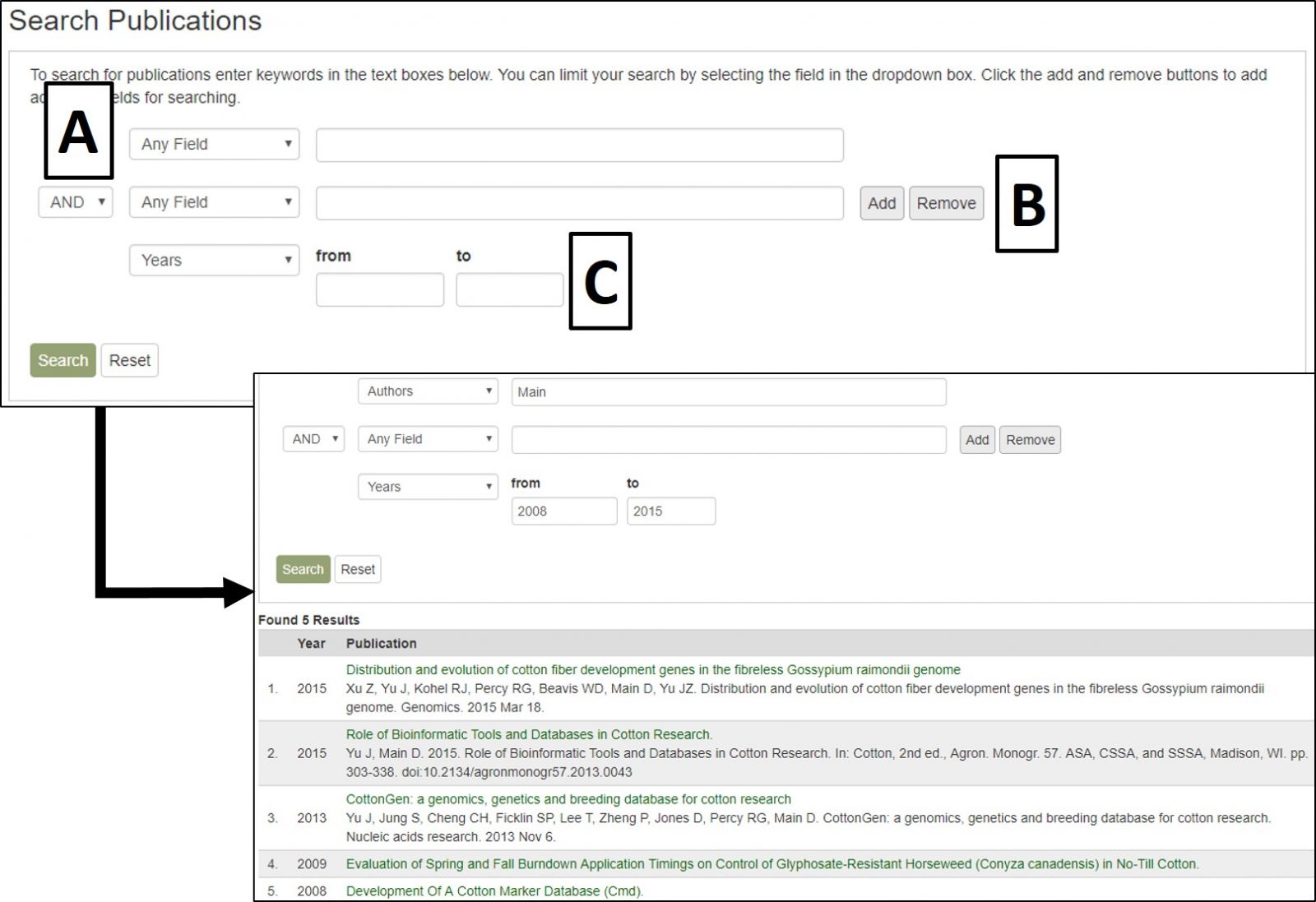
CottonGen Publication Search interface.
The search results table has information about each publication (Fig. 20A). By clicking the publication title, more detailed information is displayed. Most publications also have a link to the publisher website or PubMed record in their titles (Fig. 20B).
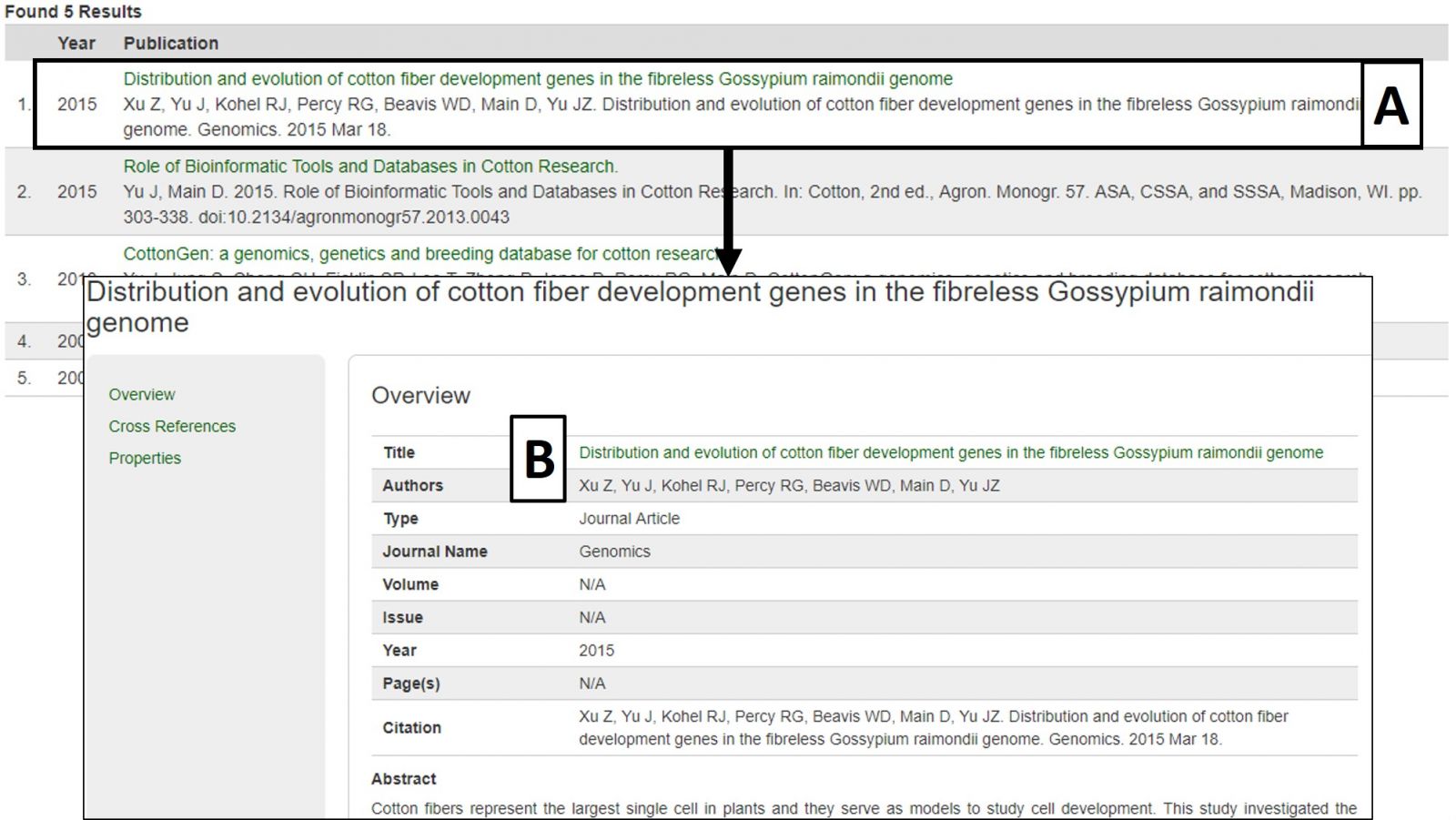
Figure 20. Publication search results and detailed publication information.
Search Markers
To search the markers in CottonGen, click on the Search menu in the header and select Search Markers. The Marker Search is useful for retrieving information about markers and can be broad or very specific depending on the parameters used. Markers can be searched individually by name or in groups by uploading a text file with multiple marker names. Filters can be set for marker type, cotton genome group, original marker species, and location (Fig. 17A). Alternatively, searches may be performed on only those markers that are mapped or are near a specific locus or QTL. Markers may also be browsed by associated genetic maps, chromosome positions, or genome groups (Fig. 17B).
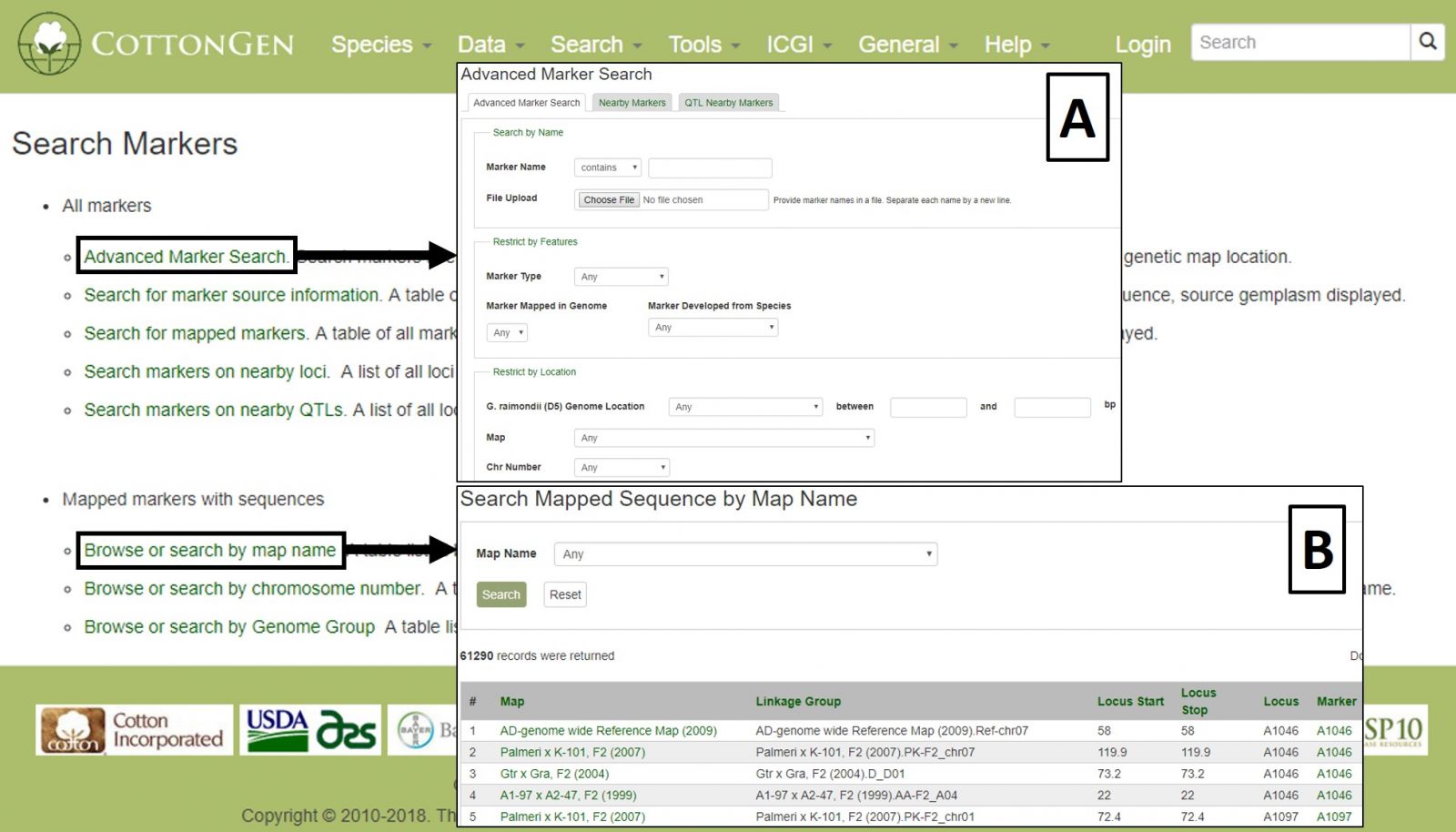
Figure 17. CottonGen Marker Search interface.
Search results are returned in table format, which is downloadable, as are associated sequences in Fasta format (Fig 18A). Clicking on a marker name displays the marker's page with information such as alignments and map positions (Fig. 18B). If you would like to change the search, either edit the parameters or click the reset button.
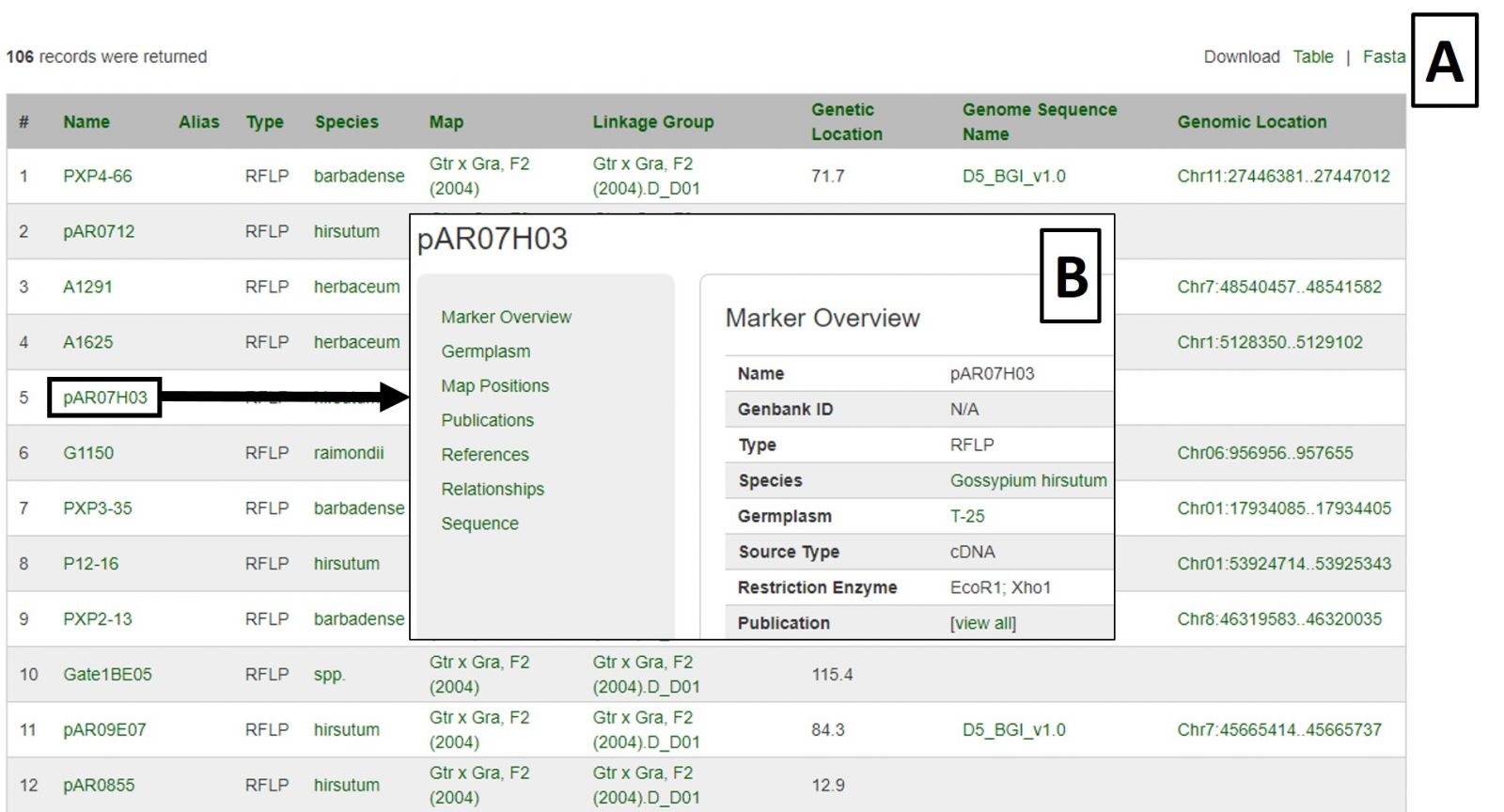
Figure 18. Marker Search results.
QTLs
Quantitative trait loci (QTLs) that are entered in CottonGen are searchable from the QTL Search option under the Search menu in the header. QTLs may be searched using trait keywords, published symbols (see the Cotton Trait Ontology list in the Data header menu), or unique identifiers assigned by CottonGen.

Figure 21. CottonGen QTL Search interface.
The results are returned in a downloadable table (Fig. 22A) with hyperlinks to more information about the QTL, the map it is located on, and the associated trait name and symbol. Clicking on the QTL label opens the overview page which has a link to map positions (Fig. 22B). From the map position table, the QTL can be viewed in MapViewer or CMap.

Figure 22. CottonGen QTL Search results.
Sequences
To search sequences, select Sequence Search from the Search header menu. This is useful for retrieving information about certain sequences from a larger dataset, or all the sequences from one or more datasets. The Gene and Transcript Search is limited to genes and transcripts, whereas the Sequence Search includes other sequence types. Use fewer filters to obtain broader results and more filters to narrow your results, as setting multiple filters will only return results matching all criteria. This is not a BLAST search; that is available under the Tools menu.
Sequence searches can be limited by species and sequence type (Fig. 23A). The search can also be restricted to a certain genome dataset and further by chromosome or scaffold location (Fig. 23B). The sequences in CottonGen can also be searched individually by name or in groups by uploading a text file with multiple names (Fig. 23C).

Figure 23. CottonGen Sequence Search interface.
Search results are returned in a table with hyperlinks to more information (Fig. 24A). The table can be downloaded, as well as a Fasta file of those sequences (Fig. 24B). A new search may be performed by changing the filters or using the Reset button.
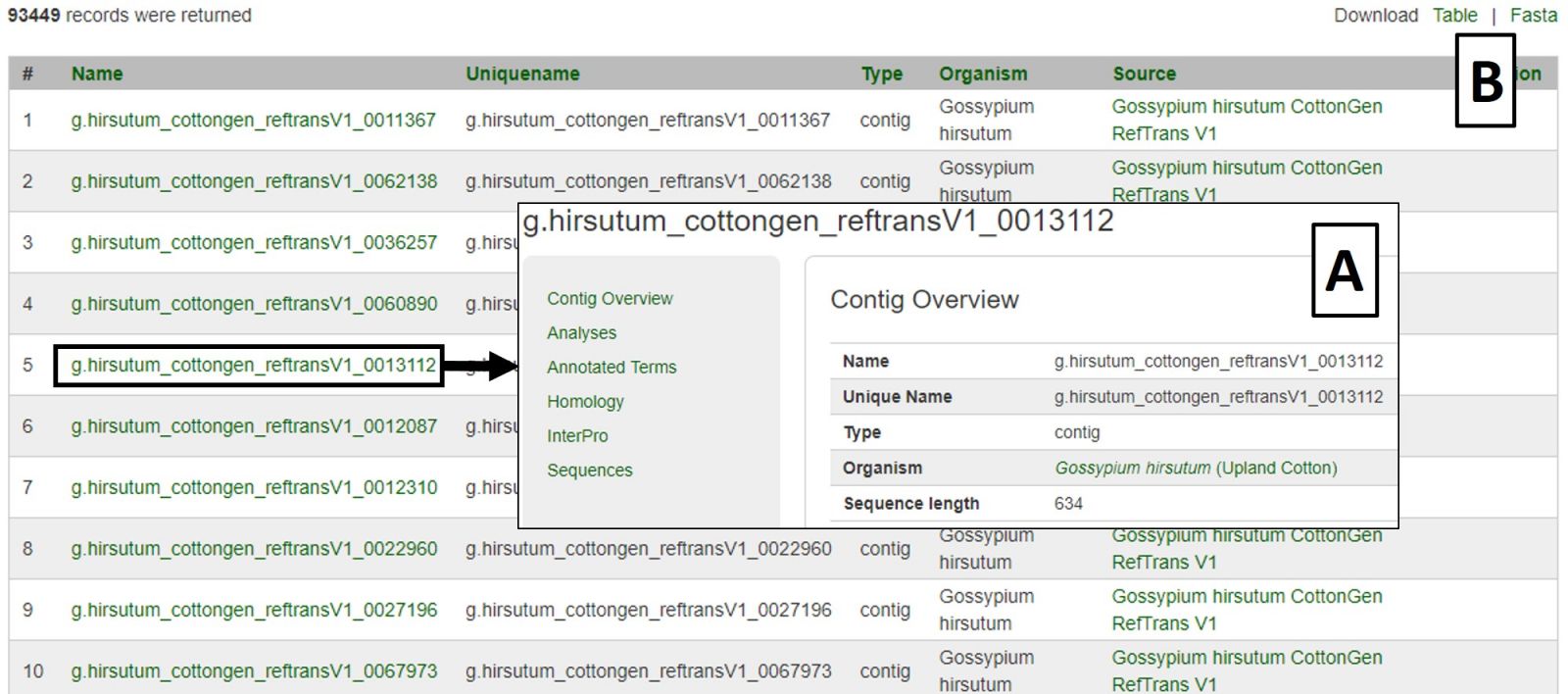
Figure 24. CottonGen Sequence Search results.
Trait Evaluations
The Trait Evaluation Search can be accessed through the Search menu in the header. This search retrieves phenotypic results from germplasm evaluation studies and is divided into Quantitative Traits (Fig. 25A) and Qualitative Traits (Fig. 25B). Quantitative Traits are those that can be measured using numerical values. Qualitative Traits are those that require the use of descriptors to evaluate.

Figure 25. CottonGen Trait Evaluation Search interface.
Quantitative Traits are searched by choosing the desired trait or traits from the pull-down menus (Fig. 26A). This will autofill the value ranges based on the minimum and maximum trait values in CottonGen and can be restricted to narrow the search (Fig. 26B). Results are displayed in a downloadable table (Fig. 26C) with links to further information on evaluated germplasms (Fig. 26D).

Figure 26. Quantitative Trait Evaluation Search interface and results.
Qualitative Traits are searched by selecting the desired trait or traits from the pull-down menus (Fig. 27A), with the option to filter using available trait descriptors once the search page has auto-updated (Fig. 27B). Results are displayed in a downloadable table (Fig. 27C) with links to further information on evaluated germplasms (Fig. 27D).

Figure 27. Qualititative Trait Evaluation Search interface and results.
Search Markers by Locus
To search the maps in CottonGen, click on the Search menu in the header and select Search Map.